Import Materials from files in various formats¶
This tutorial explains how to import materials from files in various formats into the Materials Designer interface. With the help of notebook that uses ASE python package to extract structural information from files in multiple formats (CIF, POSCAR, etc., as supported by ASE). Some formats, like espresso-in and espresso-out can be inferred from the file content.
Step 0: Open Materials Designer¶
Start by opening an instance of the Materials Designer Interface for creating and designing new Materials structures on our platform.
Step 1: Open JupyterLite Environment¶
Open the JupyterLite Environment by navigating to "Advanced" > "JupyterLite Transformation" menu item in the Materials Designer interface.
Step 2: Open the Notebook¶
Open the "Materials import from files in ASE-supported formats" in the Introduction.ipynb notebook.
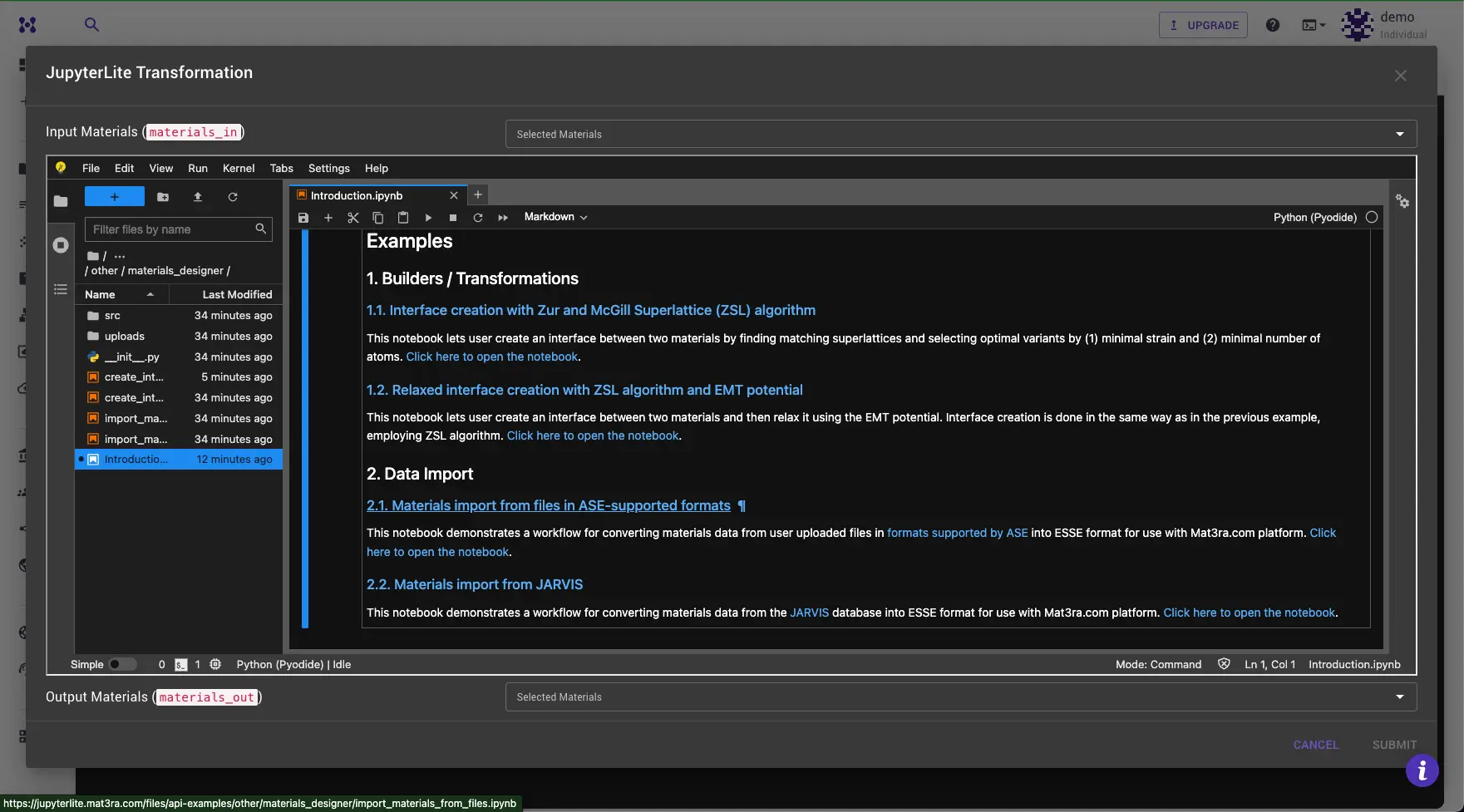
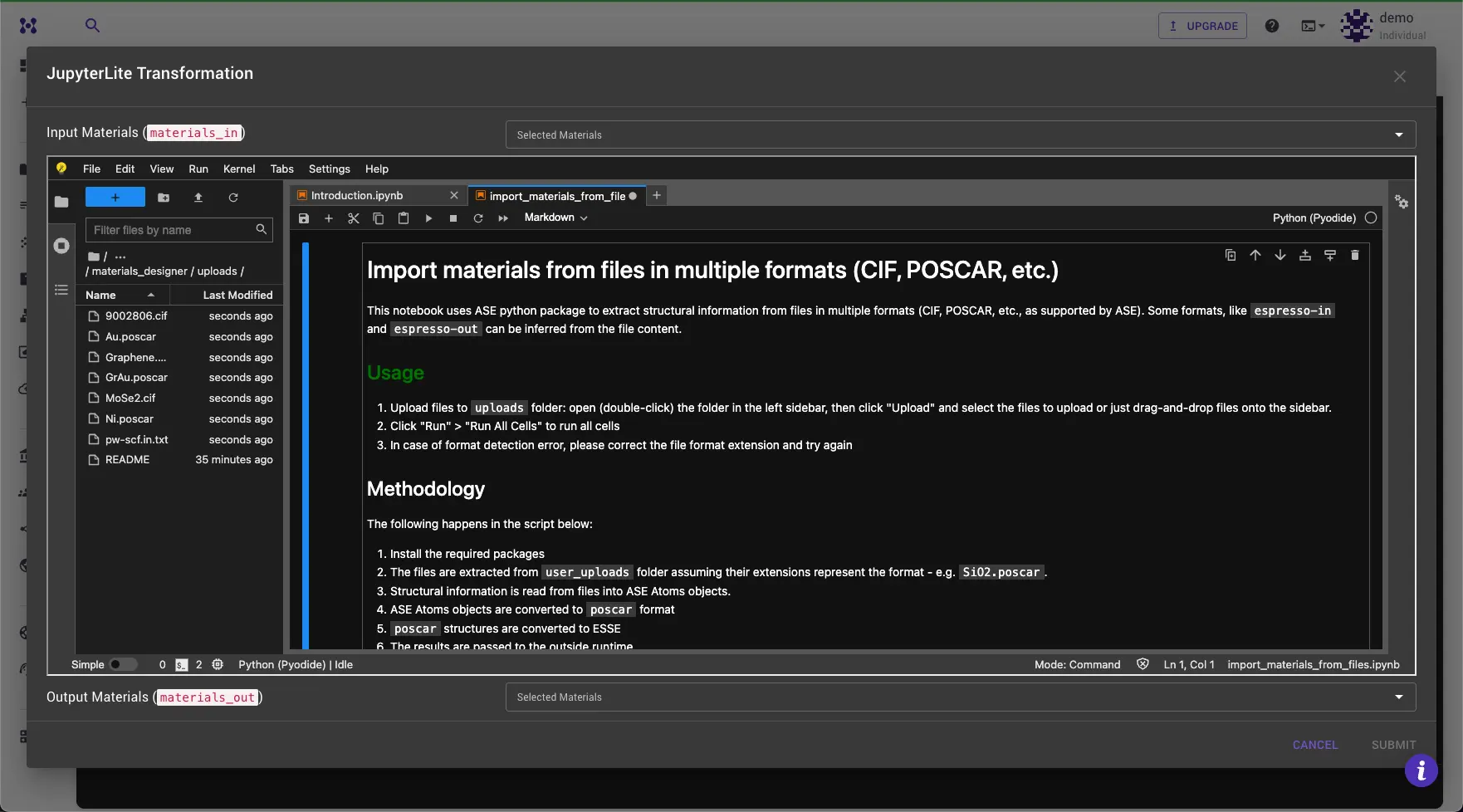
Step 3: Upload files¶
Double-click the uploads folder in the File Browser tab on the left to open it. Drag and drop the files you want to import into the field.
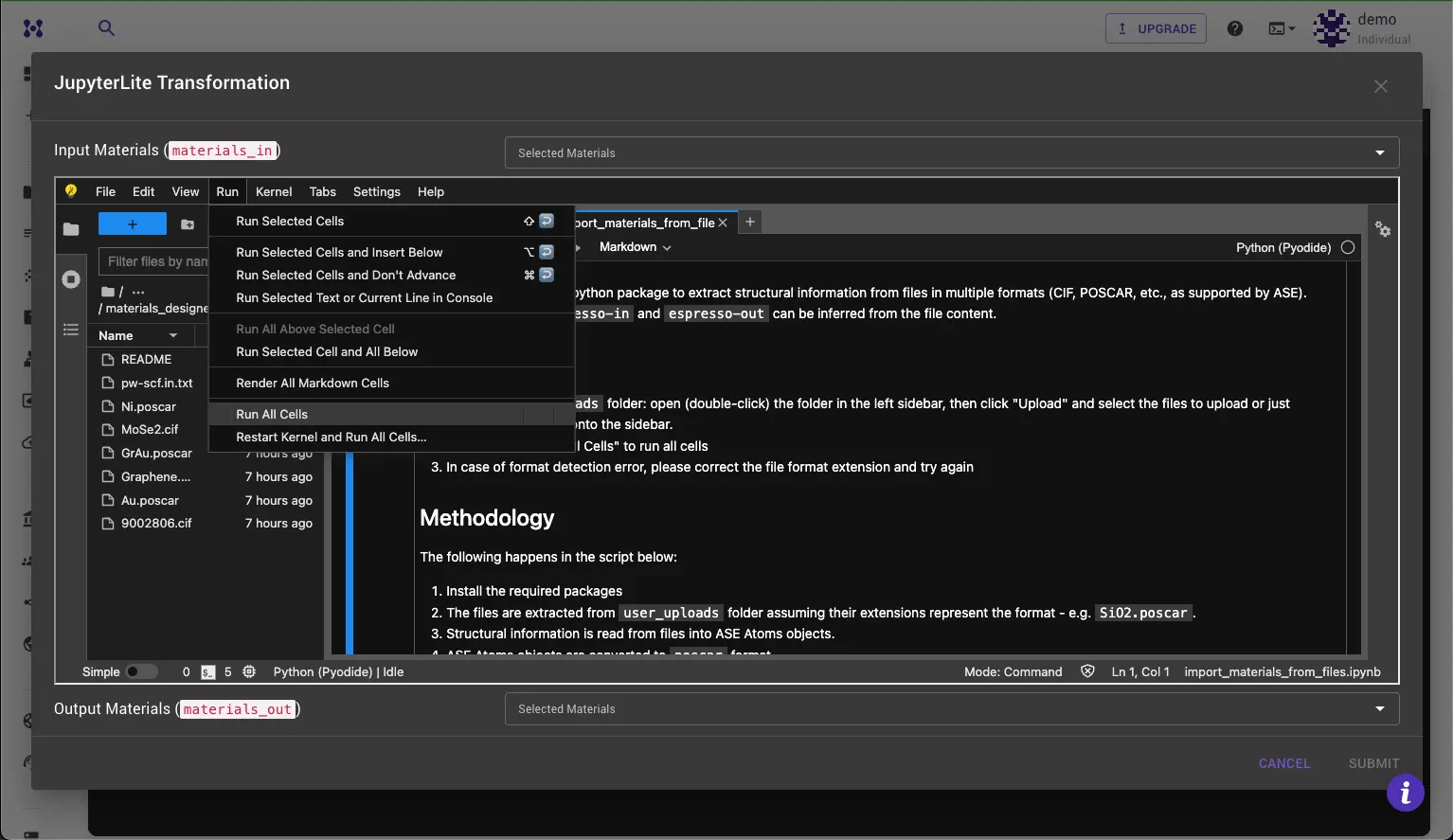
Step 4: Run the Notebook¶
Run the notebook by clicking the "Run All Cells" button in the toolbar or execute each cell by pressing "Shift + Enter" if you want to review results or change the code in the process.
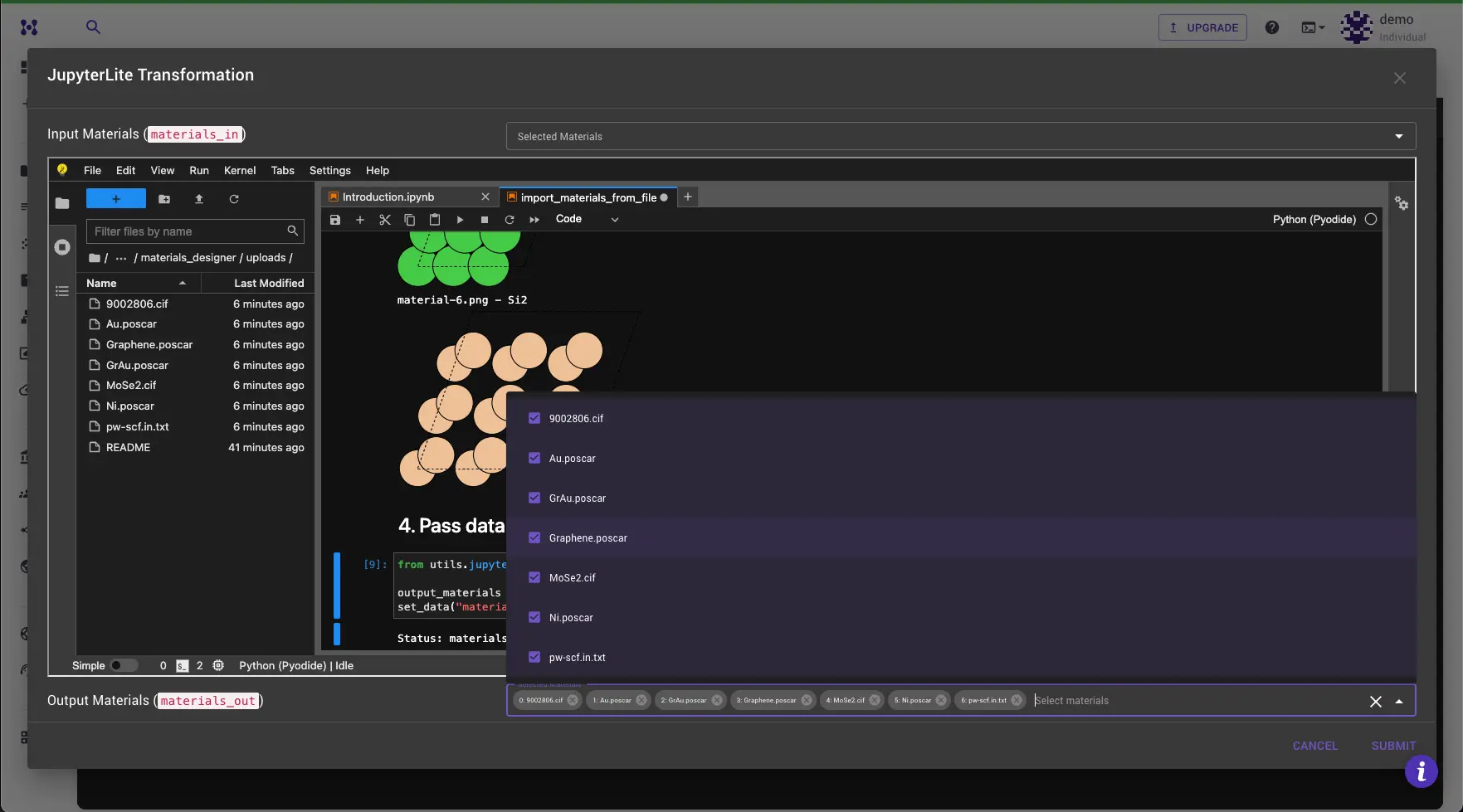
Step 5: Review the Results and Submit¶
Materials should appear in the "Materials Out" dropdown at the bottom of the dialog. Select the material you want to work with and click "Submit" to load it into the Materials Designer.
In case ASE is unable to read the file, an error message will be printed stating the unreadable files and a table of available formats. In this case, you can try to fix the issue and re-run the notebook. The error with some files does not prevent other files from being read.
Additional Information¶
ASE¶
The information about ASE IO can be found here. The version of ASE used in the JupyterLite environment is 3.22.1 (as of 2024-04-05).
Supported Formats¶
ase.io.formats.ioformats provides the list of supported formats:
| Format Name | File Extensions | Description |
|---|---|---|
| abinit-in | [] | ABINIT input file |
| abinit-out | [] | ABINIT output file |
| aims | ['in'] | FHI-aims geometry file |
| aims-output | [] | FHI-aims output |
| bundletrajectory | [] | ASE bundle trajectory |
| castep-castep | ['castep'] | CASTEP output file |
| castep-cell | ['cell'] | CASTEP geom file |
| castep-geom | ['geom'] | CASTEP trajectory file |
| castep-md | ['md'] | CASTEP molecular dynamics file |
| castep-phonon | ['phonon'] | CASTEP phonon file |
| cfg | [] | AtomEye configuration |
| cif | ['cif'] | CIF-file |
| cmdft | [] | CMDFT-file |
| cml | ['cml'] | Chemical json file |
| cp2k-dcd | ['dcd'] | CP2K DCD file |
| cp2k-restart | ['restart'] | CP2K restart file |
| crystal | ['f34', '34'] | Crystal fort.34 format |
| cube | ['cube'] | CUBE file |
| dacapo-text | [] | Dacapo text output |
| db | [] | ASE SQLite database file |
| dftb | [] | DftbPlus input file |
| dlp4 | ['config'] | DL_POLY_4 CONFIG file |
| dlp-history | [] | DL_POLY HISTORY file |
| dmol-arc | [] | DMol3 arc file |
| dmol-car | ['car'] | DMol3 structure file |
| dmol-incoor | [] | DMol3 structure file |
| elk | [] | ELK atoms definition from GEOMETRY.OUT |
| elk-in | [] | ELK input file |
| eon | ['con'] | EON CON file |
| eps | [] | Encapsulated Postscript |
| espresso-in | ['pwi'] | Quantum espresso in file |
| espresso-out | ['out', 'pwo'] | Quantum espresso out file |
| exciting | [] | exciting input |
| extxyz | ['xyz'] | Extended XYZ file |
| findsym | [] | FINDSYM-format |
| gamess-us-out | [] | GAMESS-US output file |
| gamess-us-in | [] | GAMESS-US input file |
| gamess-us-punch | ['dat'] | GAMESS-US punchcard file |
| gaussian-in | ['com', 'gjf'] | Gaussian com (input) file |
| gaussian-out | ['log'] | Gaussian output file |
| acemolecule-out | [] | ACE output file |
| acemolecule-input | [] | ACE input file |
| gen | [] | DFTBPlus GEN format |
| gif | [] | Graphics interchange format |
| gpaw-out | [] | GPAW text output |
| gpumd | [] | GPUMD input file |
| gpw | [] | GPAW restart-file |
| gromacs | ['gro'] | Gromacs coordinates |
| gromos | ['g96'] | Gromos96 geometry file |
| html | [] | X3DOM HTML |
| json | ['json'] | ASE JSON database file |
| jsv | [] | JSV file format |
| lammps-dump-text | [] | LAMMPS text dump file |
| lammps-dump-binary | [] | LAMMPS binary dump file |
| lammps-data | [] | LAMMPS data file |
| magres | [] | MAGRES ab initio NMR data file |
| mol | [] | MDL Molfile |
| mp4 | [] | MP4 animation |
| mustem | ['xtl'] | muSTEM xtl file |
| mysql | [] | ASE MySQL database file |
| netcdftrajectory | [] | AMBER NetCDF trajectory file |
| nomad-json | ['nomad-json'] | JSON from Nomad archive |
| nwchem-in | ['nwi'] | NWChem input file |
| nwchem-out | ['nwo'] | NWChem output file |
| octopus-in | [] | Octopus input file |
| proteindatabank | ['pdb'] | Protein Data Bank |
| png | [] | Portable Network Graphics |
| postgresql | [] | ASE PostgreSQL database file |
| pov | [] | Persistance of Vision |
| prismatic | [] | prismatic and computem XYZ-file |
| py | [] | Python file |
| sys | [] | qball sys file |
| qbox | [] | QBOX output file |
| res | ['shelx'] | SHELX format |
| rmc6f | ['rmc6f'] | RMCProfile |
| sdf | [] | SDF format |
| siesta-xv | [] | Siesta .XV file |
| struct | [] | WIEN2k structure file |
| struct_out | [] | SIESTA STRUCT file |
| traj | ['traj'] | ASE trajectory |
| turbomole | [] | TURBOMOLE coord file |
| turbomole-gradient | [] | TURBOMOLE gradient file |
| v-sim | ['ascii'] | V_Sim ascii file |
| vasp | ['poscar'] | VASP POSCAR/CONTCAR |
| vasp-out | [] | VASP OUTCAR file |
| vasp-xdatcar | [] | VASP XDATCAR file |
| vasp-xml | [] | VASP vasprun.xml file |
| vti | [] | VTK XML Image Data |
| vtu | ['vtu'] | VTK XML Unstructured Grid |
| wout | [] | Wannier90 output |
| x3d | [] | X3D |
| xsd | [] | Materials Studio file |
| xsf | [] | XCrySDen Structure File |
| xtd | [] | Materials Studio file |
| xyz | [] | XYZ-file |