Distributed memory calculations¶
Date: 2016/05
Overview¶
The purpose of this study was to estimate the extent to which a calculation for a single material can be efficiently scaled.
Hardware configuration
Amazon Web Services with the hardware configuration explained here were used for benchmarking. Lowest latency Ethernet network interconnect option was chosen.
Model and Method¶
Vienna Ab-initio Simulation Package (VASP) at version 5.3.5 with a corresponding set of atomic pseudo-potentials, and Quantum ESPRESSO (QE) at version 5.2.1 with a set of pseudo-potentials as explained below were employed for this study.
Two basic parallelization schemes were attempted:
- parallelization over the electronic bands for large-unit-cell materials (further referred to as ELB)
- parallelization over the sampling points in reciprocal space, or k-points (further referred to as KPT)
Compute nodes with a total of 36 CPU per node were used. Number of cores per node (PPN) and total number of nodes (NODES) were used to distinguish between parallelization levels.
Results¶
VASP-ELB¶
Inputs¶
**INCAR**
ALGO = Normal
EDIFF = 0.0001
ENCUT = 520
ISIF = 3
ISMEAR = 0
SIGMA = 0.05
ISPIN = 1
LREAL = Auto
NELM = 10
PREC = Low
# Parallelism
NCORE = 1
LPLANE = .TRUE.
**POSCAR**
50 Bi30 O108
1.0
10.866266 0.000000 -0.954010
0.000000 6.451345 0.000000
0.000000 0.000000 51.985082
Ba Bi O
50 30 108
direct
0.087932 0.000000 0.565368 Ba
0.625404 0.500000 0.646400 Ba
0.955413 0.000000 0.425732 Ba
0.340186 0.000000 0.950909 Ba
0.153840 0.500000 0.469477 Ba
0.259159 0.500000 0.126029 Ba
0.681472 0.000000 0.981631 Ba
0.125404 0.000000 0.646400 Ba
0.794831 0.500000 0.440241 Ba
0.516546 0.000000 0.318523 Ba
0.076763 0.000000 0.076531 Ba
0.949383 0.500000 0.174093 Ba
0.995789 0.000000 0.931988 Ba
0.333768 0.000000 0.512221 Ba
0.137082 0.000000 0.222918 Ba
0.653840 0.000000 0.469477 Ba
0.688638 0.500000 0.723868 Ba
0.592917 0.000000 0.821706 Ba
0.449383 0.000000 0.174093 Ba
0.385441 0.000000 0.029138 Ba
0.637082 0.500000 0.222918 Ba
0.220533 0.000000 0.365614 Ba
0.887847 0.000000 0.773393 Ba
0.720533 0.500000 0.365614 Ba
0.789340 0.000000 0.614796 Ba
0.495789 0.500000 0.931988 Ba
0.181472 0.500000 0.981631 Ba
0.576763 0.500000 0.076531 Ba
0.527676 0.000000 0.747544 Ba
0.824291 0.000000 0.272381 Ba
0.455413 0.500000 0.425732 Ba
0.980401 0.500000 0.668320 Ba
0.027676 0.500000 0.747544 Ba
0.294831 0.000000 0.440241 Ba
0.840186 0.500000 0.950909 Ba
0.833768 0.500000 0.512221 Ba
0.927808 0.000000 0.854454 Ba
0.759159 0.000000 0.126029 Ba
0.092917 0.500000 0.821706 Ba
0.885441 0.500000 0.029138 Ba
0.427808 0.500000 0.854454 Ba
0.781730 0.500000 0.873251 Ba
0.480401 0.000000 0.668320 Ba
0.324291 0.500000 0.272381 Ba
0.281730 0.000000 0.873251 Ba
0.289340 0.500000 0.614796 Ba
0.188638 0.000000 0.723868 Ba
0.016546 0.500000 0.318523 Ba
0.587932 0.500000 0.565368 Ba
0.387847 0.500000 0.773393 Ba
0.057887 0.500000 0.395436 Bi
0.104173 0.000000 0.150024 Bi
0.604173 0.500000 0.150024 Bi
0.796407 0.000000 0.199378 Bi
0.225563 0.500000 0.052079 Bi
0.711422 0.000000 0.545284 Bi
0.112287 0.500000 0.903014 Bi
0.362976 0.500000 0.349747 Bi
0.296407 0.500000 0.199378 Bi
0.987680 0.000000 0.497559 Bi
0.315493 0.500000 0.697781 Bi
0.487680 0.500000 0.497559 Bi
0.922161 0.500000 0.594720 Bi
0.219179 0.000000 0.800001 Bi
0.540108 0.500000 0.002942 Bi
0.416930 0.000000 0.101077 Bi
0.484071 0.000000 0.248330 Bi
0.984071 0.500000 0.248330 Bi
0.725563 0.000000 0.052079 Bi
0.916930 0.500000 0.101077 Bi
0.422161 0.000000 0.594720 Bi
0.815493 0.000000 0.697781 Bi
0.557887 0.000000 0.395436 Bi
0.612287 0.000000 0.903014 Bi
0.040108 0.000000 0.002942 Bi
0.719179 0.500000 0.800001 Bi
0.676341 0.500000 0.298016 Bi
0.862976 0.000000 0.349747 Bi
0.176341 0.000000 0.298016 Bi
0.211422 0.500000 0.545284 Bi
0.478903 0.264076 0.975105 O
0.794009 0.743545 0.574401 O
0.433687 0.500000 0.670087 O
0.969858 0.279329 0.892025 O
0.179468 0.726829 0.684315 O
0.580036 0.241791 0.275058 O
0.501386 0.243805 0.126414 O
0.478349 0.764772 0.625843 O
0.379555 0.276752 0.725560 O
0.824942 0.500000 0.768163 O
0.215931 0.000000 0.987806 O
0.693680 0.229357 0.927680 O
0.134064 0.729656 0.424188 O
0.203222 0.252815 0.174870 O
0.325390 0.000000 0.271903 O
0.304355 0.761555 0.828560 O
0.978349 0.264772 0.625843 O
0.105383 0.752738 0.029752 O
0.080036 0.741791 0.275058 O
0.168673 0.500000 0.363758 O
0.707662 0.000000 0.871018 O
0.304355 0.238445 0.828560 O
0.586903 0.000000 0.575600 O
0.469858 0.779329 0.892025 O
0.969858 0.720671 0.892025 O
0.879555 0.776752 0.725560 O
0.679468 0.773171 0.684315 O
0.879555 0.223248 0.725560 O
0.634064 0.770344 0.424188 O
0.469858 0.220671 0.892025 O
0.578142 0.259276 0.783096 O
0.328452 0.760817 0.075220 O
0.804355 0.261555 0.828560 O
0.379555 0.723248 0.725560 O
0.203222 0.747185 0.174870 O
0.794009 0.256455 0.574401 O
0.416818 0.257504 0.380306 O
0.294009 0.243545 0.574401 O
0.033750 0.000000 0.329333 O
0.679468 0.226829 0.684315 O
0.634064 0.229656 0.424188 O
0.102926 0.747133 0.524447 O
0.602926 0.752867 0.524447 O
0.102926 0.252867 0.524447 O
0.578142 0.740724 0.783096 O
0.828452 0.260817 0.075220 O
0.894263 0.755816 0.223836 O
0.602926 0.247133 0.524447 O
0.381754 0.263004 0.473798 O
0.792543 0.254627 0.323251 O
0.580036 0.758209 0.275058 O
0.916818 0.757504 0.380306 O
0.394263 0.255816 0.223836 O
0.193680 0.270643 0.927680 O
0.365823 0.500000 0.020410 O
0.394263 0.744184 0.223836 O
0.001386 0.256195 0.126414 O
0.634722 0.500000 0.474407 O
0.478903 0.735924 0.975105 O
0.078142 0.240724 0.783096 O
0.080036 0.258209 0.275058 O
0.292543 0.754627 0.323251 O
0.416818 0.742496 0.380306 O
0.082298 0.500000 0.080658 O
0.207662 0.500000 0.871018 O
0.645801 0.000000 0.224672 O
0.828452 0.739183 0.075220 O
0.605383 0.252738 0.029752 O
0.256293 0.000000 0.123311 O
0.134064 0.270344 0.424188 O
0.978903 0.235924 0.975105 O
0.086903 0.500000 0.575600 O
0.478349 0.235228 0.625843 O
0.693680 0.770643 0.927680 O
0.881754 0.236996 0.473798 O
0.605383 0.747262 0.029752 O
0.978903 0.764076 0.975105 O
0.328452 0.239183 0.075220 O
0.865823 0.000000 0.020410 O
0.145801 0.500000 0.224672 O
0.105383 0.247262 0.029752 O
0.294009 0.756455 0.574401 O
0.533750 0.500000 0.329333 O
0.804355 0.738445 0.828560 O
0.582298 0.000000 0.080658 O
0.179468 0.273171 0.684315 O
0.078142 0.759276 0.783096 O
0.451725 0.500000 0.175704 O
0.292543 0.245373 0.323251 O
0.703222 0.247185 0.174870 O
0.881754 0.763004 0.473798 O
0.792543 0.745373 0.323251 O
0.134722 0.000000 0.474407 O
0.756293 0.500000 0.123311 O
0.703222 0.752815 0.174870 O
0.381754 0.736996 0.473798 O
0.715931 0.500000 0.987806 O
0.894263 0.244184 0.223836 O
0.916818 0.242496 0.380306 O
0.933687 0.000000 0.670087 O
0.324942 0.000000 0.768163 O
0.193680 0.729357 0.927680 O
0.501386 0.756195 0.126414 O
0.825390 0.500000 0.271903 O
0.668673 0.000000 0.363758 O
0.978349 0.735228 0.625843 O
0.001386 0.743805 0.126414 O
0.951725 0.000000 0.175704 O
**KPOINTS**
0
Gamma
1 1 2
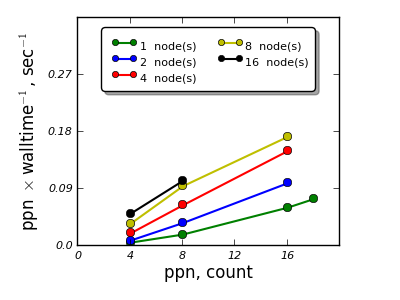
Material¶
"Ba25 Bi15 O54" with a supercell containing 188 atoms
VASP-KPT¶
Inputs¶
**INCAR**
ALGO = Normal
EDIFF = 0.0001
ENCUT = 520
IBRION = 2
ICHARG = 1
ISIF = 3
ISMEAR = 1
ISPIN = 2
LORBIT = 11
LREAL = Auto
LWAVE = False
MAGMOM = 8*0.6 16*5
NELM = 100
NCORE = 1
KPAR = <Number of compute nodes>
NSW = 1
PREC = Med
SIGMA = 0.2
**POSCAR**
Li8 V8 Mo8
1.0
11.788818 3.929606 -3.929606
-11.788818 3.929606 -3.929606
0.000000 1.964803 1.964803
Li V Mo
8 8 8
direct
0.666667 0.333333 1.000000 Li
0.958333 0.791667 0.500000 Li
0.500000 0.500000 1.000000 Li
0.208333 0.041667 0.500000 Li
0.583333 0.916667 1.000000 Li
0.333333 0.666667 1.000000 Li
0.291667 0.458333 0.500000 Li
0.125000 0.625000 0.500000 Li
0.916667 0.583333 1.000000 V
0.875000 0.375000 0.500000 V
0.625000 0.125000 0.500000 V
0.750000 0.750000 1.000000 V
0.458333 0.291667 0.500000 V
0.791667 0.958333 0.500000 V
0.083333 0.416667 1.000000 V
0.375000 0.875000 0.500000 V
0.833333 0.166667 1.000000 Mo
0.416667 0.083333 1.000000 Mo
0.708333 0.541667 0.500000 Mo
0.250000 0.250000 1.000000 Mo
1.000000 1.000000 1.000000 Mo
0.541667 0.708333 0.500000 Mo
0.041667 0.208333 0.500000 Mo
0.166667 0.833333 1.000000 Mo
**KPOINTS**
0
Gamma
6 6 6
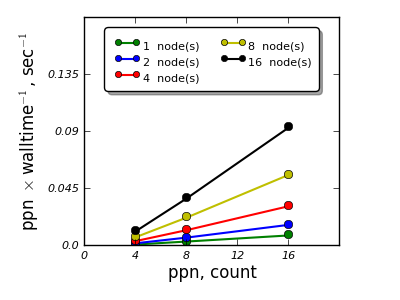
Material¶
"Li8 V8 Mo8" with a unit cell containing 24 atoms
QE-ELB¶
Inputs¶
**pw_scf.in**
&CONTROL
title = ' DEISA pw benchmark ',
calculation = 'scf',
restart_mode = 'from_scratch', ! 'restart',
tprnfor = .TRUE.,
etot_conv_thr = 1.d-5,
prefix = 'ausurf'
pseudo_dir = './'
outdir = './out/'
/
&SYSTEM
ibrav = 8,
celldm(1) = 38.7583,
celldm(2) = 0.494393,
celldm(3) = 1.569966,
nat = 112,
ntyp = 1,
nbnd = 800,
ecutwfc = 10.0,
ecutrho = 100.0,
occupations='smearing', smearing='marzari-vanderbilt', degauss=0.05
/
&ELECTRONS
diagonalization='david'
mixing_beta = 0.7
/
&IONS
ion_dynamics = 'none',
/
&CELL
cell_dynamics = 'none',
/
ATOMIC_SPECIES
AU 196.96 Au.pbe-nd-van.UPF
K_POINTS (automatic)
2 2 1 1 1 0
ATOMIC_POSITIONS (angstrom)
AU 29.285000 40.578999 7.173000
AU 29.285000 35.511002 7.173000
AU 32.214001 40.578999 7.173000
AU 32.214001 35.511002 7.173000
AU 35.141998 40.578999 7.173000
AU 35.141998 35.511002 7.173000
AU 38.070999 40.578999 7.173000
AU 38.070999 35.511002 7.173000
AU 40.999001 40.578999 7.173000
AU 40.999001 35.511002 7.173000
AU 43.928001 40.578999 7.173000
AU 43.928001 35.511002 7.173000
AU 46.855999 40.578999 7.173000
AU 46.855999 35.511002 7.173000
AU 29.285000 42.270000 4.782000
AU 29.285000 37.202000 4.782000
AU 32.214001 42.270000 4.782000
AU 32.214001 37.202000 4.782000
AU 35.141998 42.270000 4.782000
AU 35.141998 37.202000 4.782000
AU 38.070999 42.270000 4.782000
AU 38.070999 37.202000 4.782000
AU 40.999001 42.270000 4.782000
AU 40.999001 37.202000 4.782000
AU 43.928001 42.270000 4.782000
AU 43.928001 37.202000 4.782000
AU 46.855999 42.270000 4.782000
AU 46.855999 37.202000 4.782000
AU 30.749001 43.115002 7.173000
AU 30.749001 38.047001 7.173000
AU 33.678001 43.115002 7.173000
AU 33.678001 38.047001 7.173000
AU 36.605999 43.115002 7.173000
AU 36.605999 38.047001 7.173000
AU 39.535000 43.115002 7.173000
AU 39.535000 38.047001 7.173000
AU 42.464001 43.115002 7.173000
AU 42.464001 38.047001 7.173000
AU 45.391998 43.115002 7.173000
AU 45.391998 38.047001 7.173000
AU 48.320999 43.115002 7.173000
AU 48.320999 38.047001 7.173000
AU 30.749001 34.666000 4.782000
AU 30.749001 39.737999 4.782000
AU 33.678001 34.666000 4.782000
AU 33.678001 39.737999 4.782000
AU 36.605999 34.666000 4.782000
AU 36.605999 39.737999 4.782000
AU 39.535000 34.666000 4.782000
AU 39.535000 39.737999 4.782000
AU 42.464001 34.666000 4.782000
AU 42.464001 39.737999 4.782000
AU 45.391998 34.666000 4.782000
AU 45.391998 39.737999 4.782000
AU 48.320999 34.666000 4.782000
AU 48.320999 39.737999 4.782000
AU 29.285000 40.578999 0.000000
AU 29.285000 35.511002 0.000000
AU 32.214001 40.578999 0.000000
AU 32.214001 35.511002 0.000000
AU 35.141998 40.578999 0.000000
AU 35.141998 35.511002 0.000000
AU 38.070999 40.578999 0.000000
AU 38.070999 35.511002 0.000000
AU 40.999001 40.578999 0.000000
AU 40.999001 35.511002 0.000000
AU 43.928001 40.578999 0.000000
AU 43.928001 35.511002 0.000000
AU 46.855999 40.578999 0.000000
AU 46.855999 35.511002 0.000000
AU 30.749001 41.424000 2.391000
AU 30.749001 36.355999 2.391000
AU 33.678001 41.424000 2.391000
AU 33.678001 36.355999 2.391000
AU 36.605999 41.424000 2.391000
AU 36.605999 36.355999 2.391000
AU 39.535000 41.424000 2.391000
AU 39.535000 36.355999 2.391000
AU 42.464001 41.424000 2.391000
AU 42.464001 36.355999 2.391000
AU 45.391998 41.424000 2.391000
AU 45.391998 36.355999 2.391000
AU 48.320999 41.424000 2.391000
AU 48.320999 36.355999 2.391000
AU 29.285000 43.959999 2.391000
AU 29.285000 38.893002 2.391000
AU 32.214001 43.959999 2.391000
AU 32.214001 38.893002 2.391000
AU 35.141998 43.959999 2.391000
AU 35.141998 38.893002 2.391000
AU 38.070999 43.959999 2.391000
AU 38.070999 38.893002 2.391000
AU 40.999001 43.959999 2.391000
AU 40.999001 38.893002 2.391000
AU 43.928001 43.959999 2.391000
AU 43.928001 38.893002 2.391000
AU 46.855999 43.959999 2.391000
AU 46.855999 38.893002 2.391000
AU 30.749001 43.115002 0.000000
AU 30.749001 38.047001 0.000000
AU 33.678001 43.115002 0.000000
AU 33.678001 38.047001 0.000000
AU 36.605999 43.115002 0.000000
AU 36.605999 38.047001 0.000000
AU 39.535000 43.115002 0.000000
AU 39.535000 38.047001 0.000000
AU 42.464001 43.115002 0.000000
AU 42.464001 38.047001 0.000000
AU 45.391998 43.115002 0.000000
AU 45.391998 38.047001 0.000000
AU 48.320999 43.115002 0.000000
AU 48.320999 38.047001 0.000000
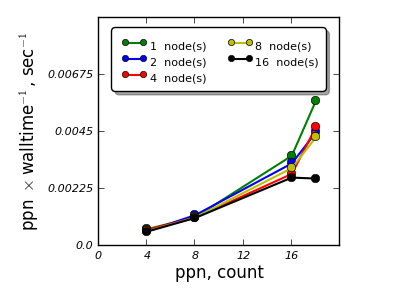
Material¶
Aluminum surface containing 112 atoms
QE-KPT¶
Inputs¶
**pw_scf.in**
&control
calculation='scf'
restart_mode='from_scratch'
wf_collect = .true.
prefix='FeSe_bs'
pseudo_dir = './'
outdir='./tmp'
tprnfor = .true.
tstress = .true.
!nosym = .true.
/
&system
ibrav = 6
celldm(1) = 7.114
celldm(3) = 5.8624
nat = 4
ntyp = 3
ecutwfc = 40
ecutrho = 400
occupations='smearing'
smearing='methfessel-paxton',
degauss=0.025
!nspin = 2
starting_magnetization(1) = 0.10
starting_magnetization(2) = -0.10
!la2F = .true.
nbnd = 45
!nosym = .true.
/
&electrons
diagonalization='cg'
mixing_beta = 0.5
conv_thr = 1.0d-10
/
&ions
ion_dynamics = 'bfgs'
/
&cell
cell_dynamics = 'bfgs'
cell_dofree = 'xy'
/
ATOMIC_SPECIES
Fe1 55.845 fe_pbe_gbrv_1.5.upf
Fe2 55.845 fe_pbe_gbrv_1.5.upf
Se 78.960 se_pbe_gbrv_1.0.upf
ATOMIC_POSITIONS crystal
Fe1 0.750000000 0.250000000 0.000000000
Se 0.250000000 0.250000000 0.063815979
Se 0.750000000 0.750000000 -0.063815979
Fe2 0.250000000 0.750000000 0.000000000
K_POINTS automatic
32 32 1 0 0 0
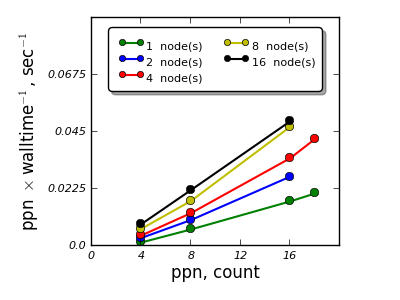
Material¶
FeSe monolayer with 4 atoms
Conclusions¶
- VASP and QE were studied for scalability for a single material - single calculation,
- K-point sampling based parallelization appears to be feasible and scales efficiently up to 16 nodes,
- Parallelization over the electronic bands for the cases studied shows efficient scalability up to 4 nodes for VASP, for QE an adjustment of parallelization parameters is necessary to reach efficient parallelization over electronic bands.